I recall back in the late 1980’s, working for Smith Kline and French, reading a couple of articles in Tet. Letts. on biotransformations. I was intrigued, and a bit sceptical, that such transformations with enzymes would take place and lead to efficient large-scale processes. I had no experience or training in this area. My degree was in chemistry, PhD in organometallics, and I worked in the Chemical Development function. I purchased a few commercially available enzymes – pig liver esterase, subtilisin protease, Candida Antarctica lipase and a few others and started screening potential reaction candidates. To my delight some worked, most did not. Typically, no reaction, poor reaction rate, not sufficient ee, or high ee but wrong enantiomer! In some cases, reaction engineering could give some improvement, but in most cases, this was a dead end. In 1989, SK&F merged with Beecham’s – a accompany steeped in a history of antibiotic development and biotransformation. I recall quickly making contact and good friends within the research biotechnology group at Brockham park and the scale up and development biotech group at Worthing, and their help and guidance encouraged me to keep going and start to scale up some biocatalysed processes.
Nevertheless, the biggest showstopper still was what to do if a commercial enzyme did not quite hit the desired product quality. Evolution and heterologous expression were known, but was lengthy, costly, and not always successful. Other options were to screen large collections of whole cell organisms to look for activity-if found, use a whole cell process or isolate the desired enzyme – again, not a quick process. Microorganisms could be sought in all kinds of bizarre places – pond sludge, used tea leaves and duck faeces being some of the most bizarre I encountered at the start of my journey into industrial biotechnology. Other options were to try and grow organisms in limited media and hope that a strain would evolve relevant to utilise your substrate as a carbon or nitrogen source. To look for strains/mutants more productive at producing enzymes or natural products like clavulanic acid, random mutation using ionising radiation or chemical mutagens like N,N-dimethyl nitrosamine would often be used – a lengthy process that could take years to reach (if ever) industrially relevant titres.
Where are we now in the field of enzyme identification and scale -up ? Well, it’s a different world – techniques and technologies exist that would have been thought science fiction back in the 1980’s.
A whole suite of tools and technologies has been developed to make identification and development of industrially useful enzymes faster, quicker and with a much higher degree of success – Figure 1.
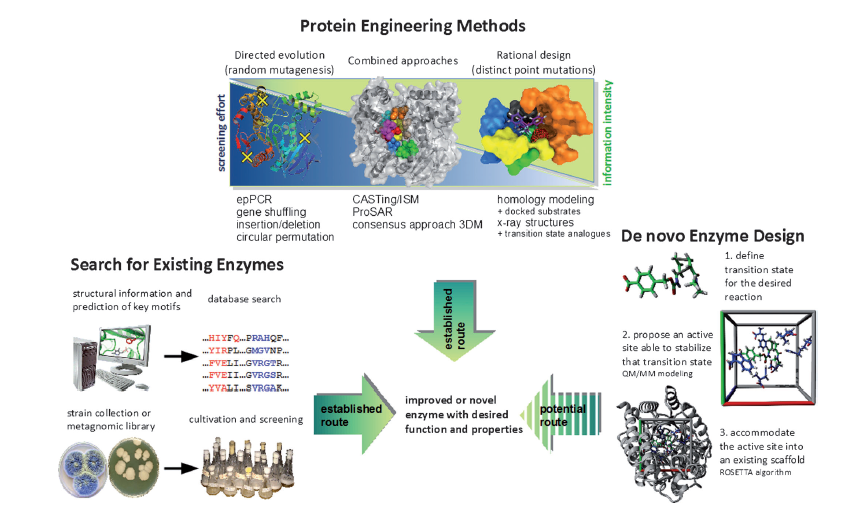
Figure 1 (Ref 1)
This has given rise to an increasing range of enzymic transformations and key enabling molecular biology techniques that have become part of the everyday toolbox of enzyme development-Figure 2.
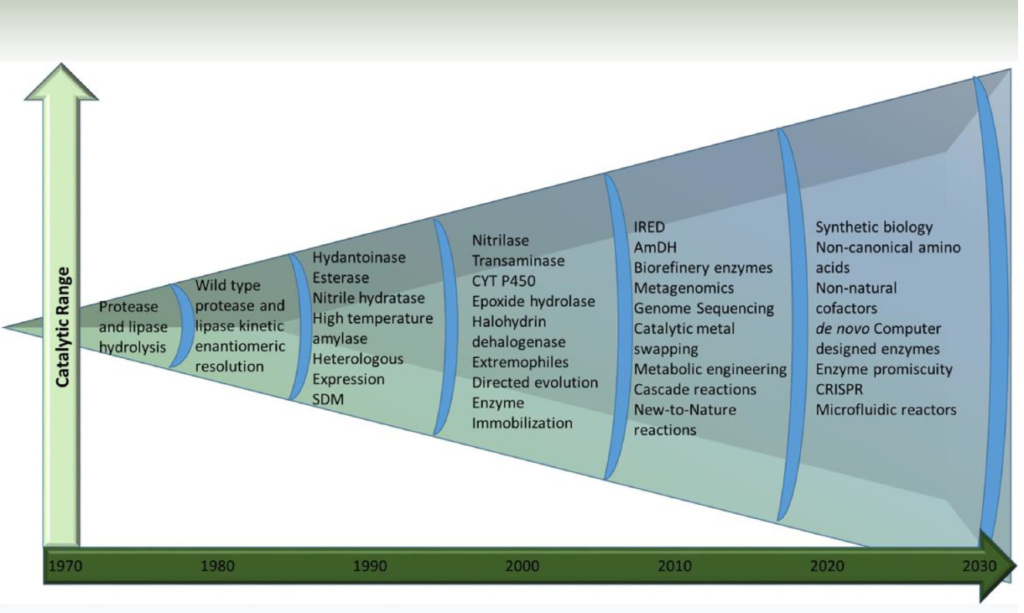
Figure 2 (Ref 2)
Maybe not currently open to all, but some extortionary exemplars of scientific achievement are the complete chemical synthesis of long DNA transcriptable in human cells (ref 3), and the chemical synthesis of mirror image proteins using D amino acids such as a 312-residue chaperone dependent DapA protein (ref 4), and an enzymatically active mirror-Image 252 residue DNA-Ligase made of D-amino acids (ref 5). Looking at just how far the use of enzymes can be advanced in the synthesis of pharmaceuticals, the Merch synthesis of Islatravir (Ref 6) is an impressive example – nine enzymes – many evolved using the modern molecular biology toolbox.
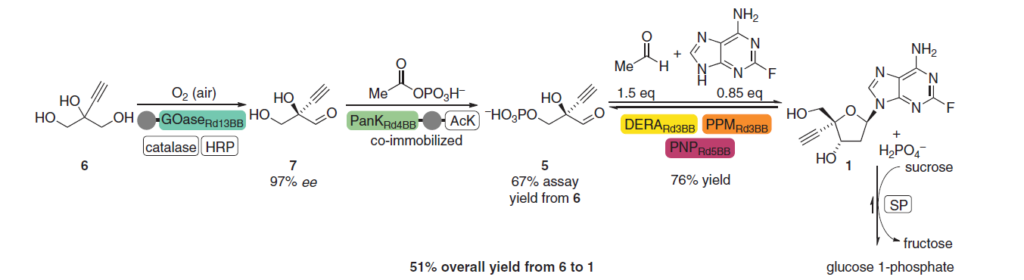

Figure 3 Merck synthesis of Islatravir (Ref 6)
Alongside modern techniques for directed evolution of mutant enzymes (Ref 7) further important advances are the ability to incorporate non-canonical or unnatural amino acids into enzyme sequences giving properties such as increased stability and active sites that can catalyse chemistry not used in nature (Ref 8). A rapidly growing area is the application of known enzymes as promiscuous biocatalysts, catalysing chemistry not seen in nature. The most famous example being the repurposing of p450 enzymes and hemes as catalysts for chiral cyclopropanation (Ref 9).
As might be expected, the application of AI/MI and in silico modelling becomes more important with the speed and accuracy of in silico models to predict enzyme structure are giving access to enzyme sequences never seen in nature and enzymes designed to catalyse reactions -Kempases, esterases, Diels-Alderases, and enzymes for Michael and Knoevenagel reactions -the list grows every year (Ref 10! The power and accuracy of the prediction developed by enterprises like Deep Mind/Alpha fold is staggering – making the in silico design of active biocatalysts as an option to maybe cut rounds of evolution needed to hit key performance parameters (Ref 11).
So, we can rapidly evolve known enzymes and design novel enzymes in silico. Where else could we find diverse and active enzymes? According to a new estimate, there are about one trillion species of microbes on Earth, and 99.999 percent of them have yet to be discovered. As recently as 1998, the number of microbial species was thought to be a few million at most — little more than the number of insect species. But estimates have been growing ever since. Of the known bacteria, only ~ 2% can be cultured – we don’t understand conditions needed for growth/symbiosis with other microorganisms. So, does this put a huge number of potentially useful enzymes out of reach? Metagenomics (environmental and community genomics) provides a route to tap into these novel enzyme sequences (Ref 12). This involves:
- Looking for environmental DNA (genetic material released from lysed cells)
- Isolate/ sequence/ amplify – look for sequences that could encode for useful activity – esterase, transaminase, ketoreductase etc.
- Clone into host microorganisms that we do know how to grow
Thirty years ago, metagenomics was seen as somewhat of a herculean task –now modern analytical techniques, rapid cheap gene sequencing and synthesis alongside the ability to look for sequence homology across millions of known enzyme structures have considerably lowered the activation barrier and greatly increased the chance of success.
References:
- Synth. Catal. 2011, 353, 2191 – 2215
- ChemSusChem 2019, 12, 2859 – 2881
- ChemBioChem 2021, 23, Pages 3273-3276
- Proc Natl Acad Sci U S A, 2014 Aug 12;111(32):11679-84.
- Cell Chemical Biology 26, 645–651, May 16, 2019
- Science 366, 1255–1259 (2019)
- Rev. 2021, 121, 20, 12384–12444
- ChembBioChem 2020 21, 2241-2249; Rev. 2021, 121, 10, 6173–6245
- ACS Catal. 2016, 6, 7810−7813; ChemBioChem 2019, 20, 1129 – 1132
- ChemBioChem 2020 21, 2241-2249
- https://www.nature.com/nmeth/volumes/19/issues/1
- Nature Biotechnology v35 No. 9 SEPTEMBER 2017 833; Microbiology and Molecular Biology Reviews, 2004, 669–685
About the Author:
Dr Andrew Wells, Charnwood Technical Consulting is a specialist and expert in the field of chemical process R&D where, over the past 30 years, he have spent most of his professional career employed at three multi-national pharmaceutical companies, rising to the rank of Senior Principal Scientist (Director Level) at AstraZeneca. He has experience across all aspects of the pharmaceutical lifecycle, especially R&D, synthesis, green chemistry and material supply at all scales from discovery chemistry through to validation, manufacture and life cycle management. A major focus over the past 20 years has been the application of homo/hetero-metal catalysis, biotransformations /biotechnology and green chemistry solutions in the tactical and strategic manufacture of pharmaceuticals, agrochemicals and fine chemicals. His career has spanned across a broad range of scientific disciplines, and he is a keen exponent of working at the interface of the physical and biological sciences to champion the adoption of novel, more sustainable technologies across a range of business areas. Andy is also a tutor on several training courses: Biocatalysis as a Tool for the Industrial Chemist, Solvents & Solvent Selection in Chemical Manufacturing and Biocatalysis – Solving Scale up Issues with Biocatalyzed Reactions.








